IN A NUTSHELL
1. About This Work
Feature extraction or feature encoding is a fundamental step in the construction of high-quality machine learning-based models. Specifically, this step is key to determining the effectiveness of trained models in bioinformatics applications (Chou, 2011). In the last two decades, a variety of feature encoding schemes have been proposed in order to exploit useful patterns from protein sequences. Such schemes are often based on sequence information or representation of physicochemical properties. Although direct features derived from sequences themselves (such as amino acid compositions, dipeptide compositions and counting of k-mers) are regarded as essential for training models, an increasing number of studies have shown that evolutionary information in the form of PSSM profiles is much more informative than sequence information alone (An, et al., 2016). Accordingly, PSSM-based feature descriptors have been commonly used as indispensable primary features to construct models, filling a major gap in the current bioinformatics research. For example, PSSM-based feature descriptors have successfully improved the prediction performance of structural and functional properties of proteins across a wide spectrum of bioinformatics applications. These include for example protein fold recognition (Lobley, et al., 2009) and the prediction of protein structural classes (Liu, et al., 2010), protein-protein interactions (Zahiri, et al., 2013), protein subcellular localization (Xie, et al., 2005), RNA-binding sites (Cheng, et al., 2008), and protein functions (Radivojac, et al., 2013), to name a few.
A number of servers and standalone software packages have been developed to derive a variety of feature descriptors from protein, DNA and RNA sequences, including PROFEAT (Rao, et al., 2011), PseAAC (Shen and Chou, 2008), propy (Cao, et al., 2013), repDNA (Liu, et al., 2015), protr/ProtrWeb (Xiao, et al., 2015), Pse-in-One (Liu, et al., 2015), repRNA (Liu, et al., 2016) and Pse-Analysis (Liu, et al., 2017). Despite their usefulness and popularity, these tools primarily focus on the generation of features related to sequence-based and/or physicochemical descriptors, instead of PSSM profile-based features. Indeed, there are over 20 different PSSM-based algorithms that calculate and model PSSM-based feature descriptors. However, to the best of our knowledge, there is currently no consolidated web server or toolkit available for generating these PSSM-based feature descriptors. Here, we present a bioinformatics toolkit, POSSUM, an effective tool that enables users to generate a broad spectrum of PSSM-based numerical representation schemes for protein sequences. It implements a wide range of algorithms available in the literature, provides an easy-to-use interface, and offers much needed functionality and flexibility for users to derive and customize these descriptors. We demonstrate the usage of POSSUM-calculated PSSM features for the prediction of bacterial secretion effector proteins.
ARCHITECTURE
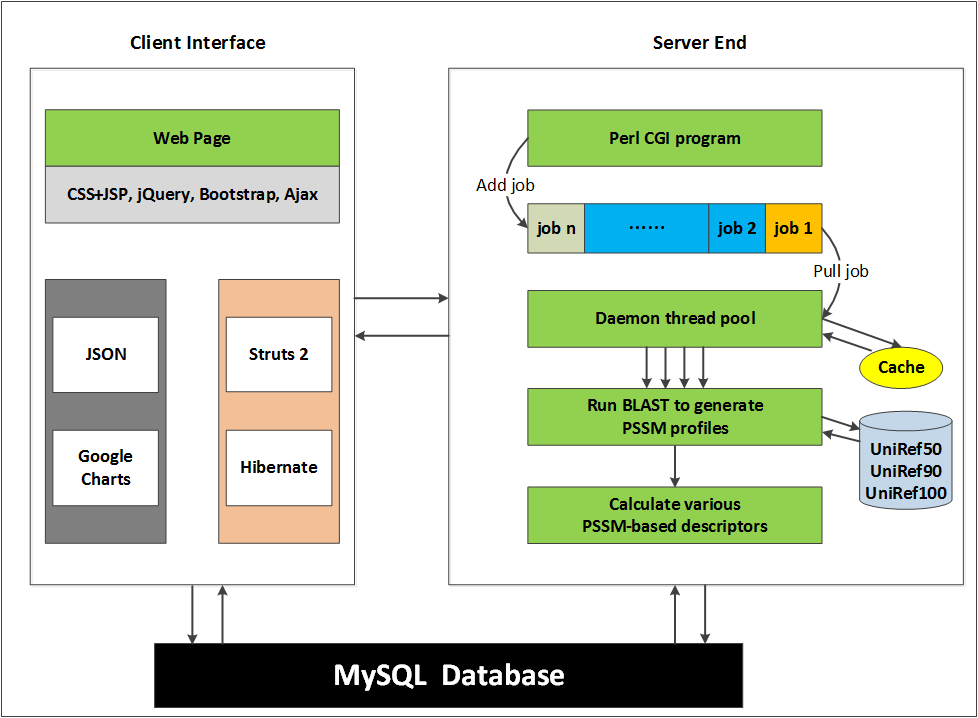
1. Architecture of the web server
The architecture of the POSSUM web server consists of two parts: client interface and server end. The client interface is implemented by Jquery, Bootstrap, Struts and Hibernate, which interact with users and forward users' submitted jobs to the server end. For the server end, Perl CGI program lines up the jobs in a queue and invokes a Perl daemon thread for each job to execute the descriptor generation process. This architecture guarantees that a number of jobs can be executed simultaneously (The max number of allowed thread is predefined in POSSUM), while the rest of jobs will queue in turn.
2. Architecture of the Toolkit
The architecture of the POSSUM standalone toolkit is displayed in the following picture. The toolkit was implemented in Python (for core function implementation) and Perl (for universal command line interface).
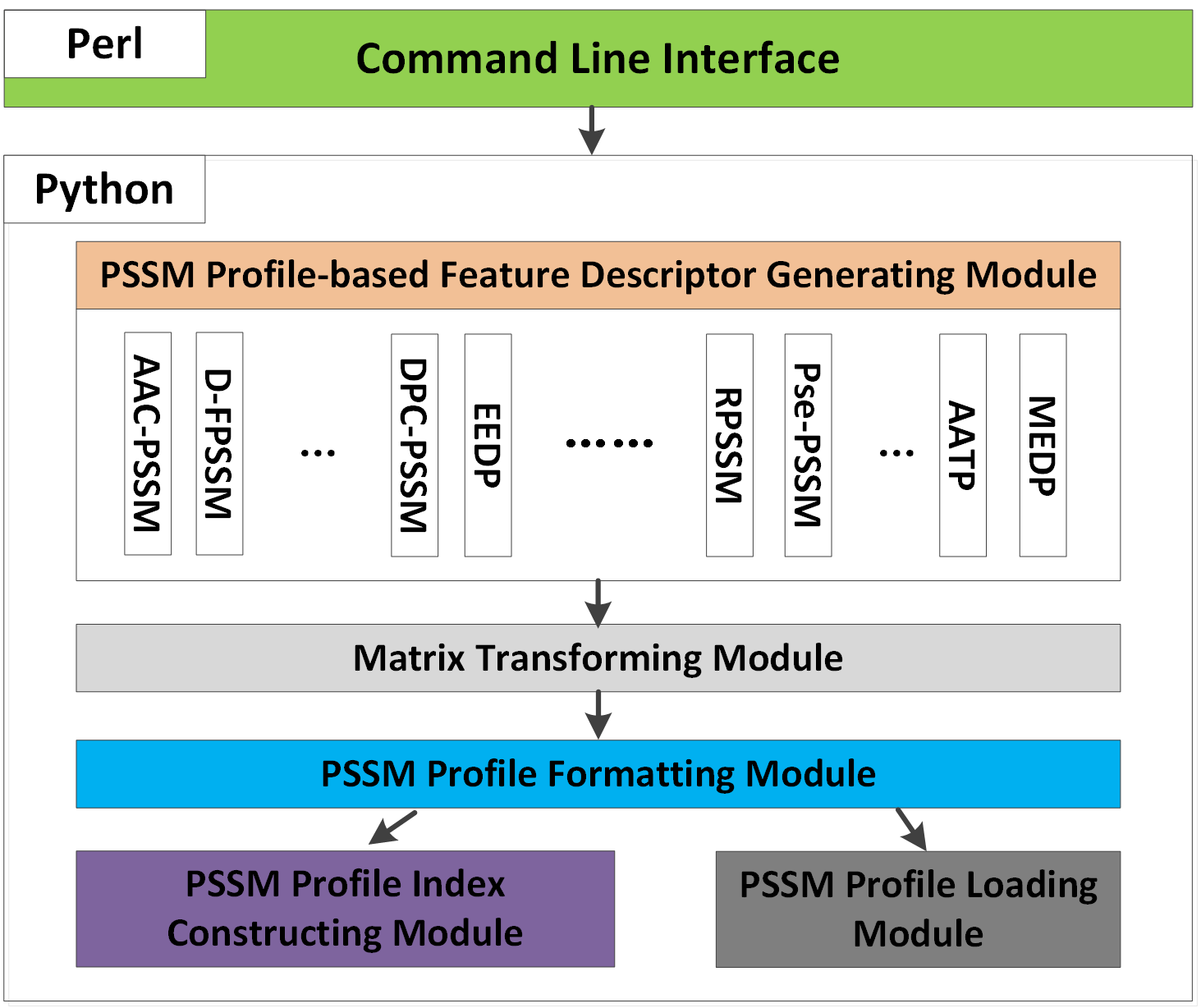
The major components of the toolkit are briefly described as follows:
-
Command Line Interface: This module is made available to provide a universal and user-friendly command line interface, via which users can effectively interact with the toolkit. This module allows users to specify and apply different parameters and it invokes the descriptor generating process. -
PSSM Profile-based Feature Descriptor Generating Module: This module can be used to wrap up and output the descriptor files based on the raw descriptor vectors (generated by the Matrix Transforming Module) in accordance with the user-specified parameters. -
Matrix Transforming Module: This module can be used to transform the PSSM matrix (which is abstracted from the original PSSM profile) to generate user-specified raw descriptor vectors. Various applicable matrix transformation functions in groups of row transformations, column transformations, and mixture of row and column transformations are available within this module. -
PSSM Profile Formatting Module: This module can be used to abstract the PSSM matrix from the PSSM profile. -
PSSM Profile Index Constructing Module: This module is a fundamental part of the program that scans the FASTA sequences and the PSSM profile folder to build a hash map for each query sequence and its corresponding PSSM profile. -
PSSM Profile Loading Module: This module looks up the hash table (built by the PSSM Profile Index Constructing Module) to check the availability of the PSSM profile for a sequence and loads the corresponding PSSM profile into the memory.
ONLINE WEB SERVER
1. Textarea Input
1.1 Input formats
Two types of input are allowed for POSSUM: sequences in FASTA format (recommended) or raw sequences.
For sequences in FASTA format, you can input as follows:
Also, the following input (which is the original formats downloaded from Uniprot database)
will be formated (without line break inside the sequence) as:
For the raw sequences, you can input as follows:
which will be formated by POSSUM as follows:
1.2 Input limits
- The length of each submitted sequence should be in the range of 50 to 5000 characters.
- Since PSSM-based feature computing is a time-consuming job, especially the process of generating PSSM, the maximum number of submitted sequences each time should be no more than 500.
- The submitted sequences should not contain illegal characters, such as "B", "J", "O", "U", "X" and "Z".
2 Upload a file in fasta format
Instead of inputing sequences directly in the textarea, users can also upload a file in fasta format.
Note: please don't submit sequences by textarea and file uploading simultaneously. Users should choose one or the other for each submission.
3 Select algorithms to generate descriptors
PSSM-based algorithms are designed based on the matrix transformation from the original PSSM, which can be categorized into three types according to the thought of matrix transformation: row transformation, column transformation, and mixture of row and column transformation. For POSSUM, these descriptors are divided into four groups. The first group consists of AAC-PSSM, D-FPSSM, smoothed-PSSM, AB-PSSM, PSSM-composition, RPM-PSSM and S-FPSSM, which are generated by row transformation of the original PSSM. The second group contains the descriptors generated by column transformation, including DPC-PSSM, k-separated-bigrams-PSSM, tri-gram-PSSM, EEDP and TPC. The third group includes EDP, RPSSM, Pse-PSSM, DP-PSSM, PSSM-AC and PSSM-CC, which are generated by mixture of row and column transformation. The fourth group comprises of AADP-PSSM, AATP and MEDP, which simply combine descriptors in the former three groups.
There are 6 algorithms that require users to input parameters, including smoothed-PSSM, k-separated-bigrams-PSSM, Pse-PSSM, DP-PSSM, PSSM-AC and PSSM-CC.
- For smoothed-PSSM, smoothing_window denotes the size of smoothing window and should be an odd number; sliding_window denotes the size of sliding window.
- For k-separated-bigrams-PSSM, k denotes the distance between the amino acid positions, with a default value of 1.
- For Pse-PSSM, ξ denotes the ξ most contiguous PSSM scores along the protein chain, with a default value of 1.
- For DP-PSSM, α denotes α-th amino acid afterward, with a default value of 5.
- For PSSM-AC and PSSM-CC, LG denotes the maximum distance of two residues along the sequence, with a default value of 10.
4 Select database for BLAST
Currently, three databases (uniref50, uniref90, uniref100) are available in POSSUM. A user can select uniref50 for BLAST to generate a PSSM in a more efficient way, or can generate more accurate PSSM files by selecting uniref90 or uniref100 at the expense of the speed.
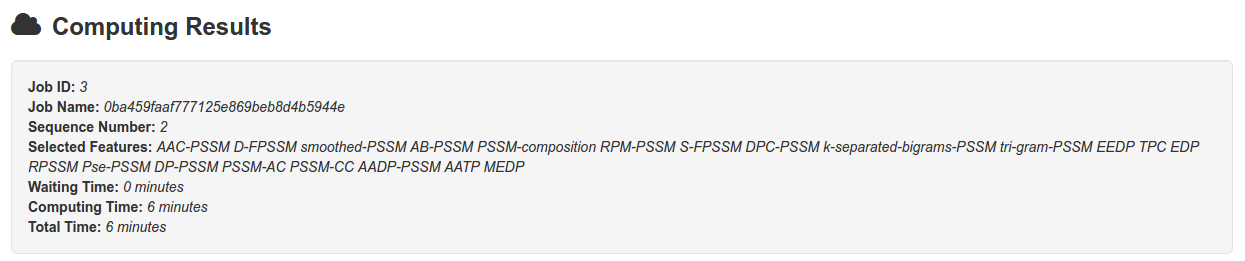
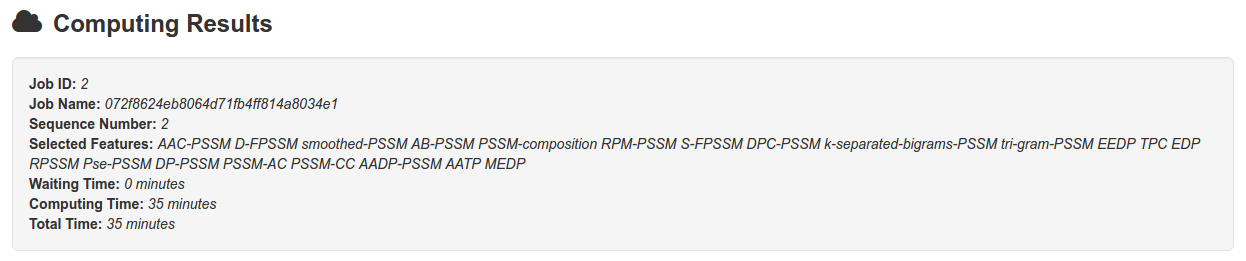
Typically, for the two sequences in example, it takes 6 minutes, 35 minutes and 71 minutes to complete the job by using uniref50, uniref90 and uniref100, respectively.

5 Output
There exist 3 types of output pages for the result of computation: result page, warning page and error page.
5.1 Result page
The result page consists of 3 parts: job information, PSSM files download and PSSM-based feature files download.
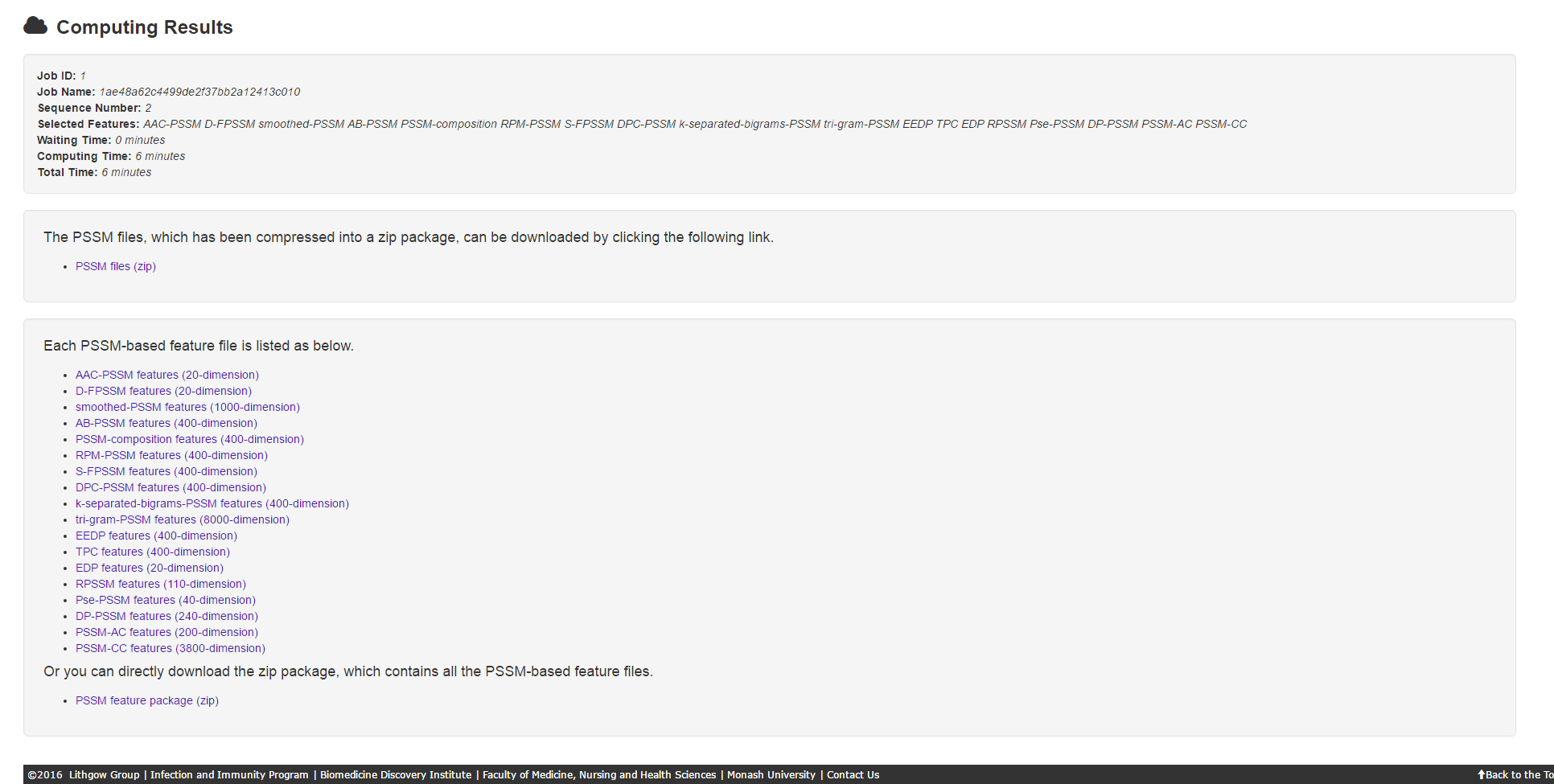
5.2 Warning page
Should any sequences be missed during the generation of PSSM files (this probability is extremely small, but is finite.). Users will receive a warning page, which contains 4 parts: job information, missed fasta sequences file, PSSM files of the rest sequences and PSSM-based feature files of the rest sequences.
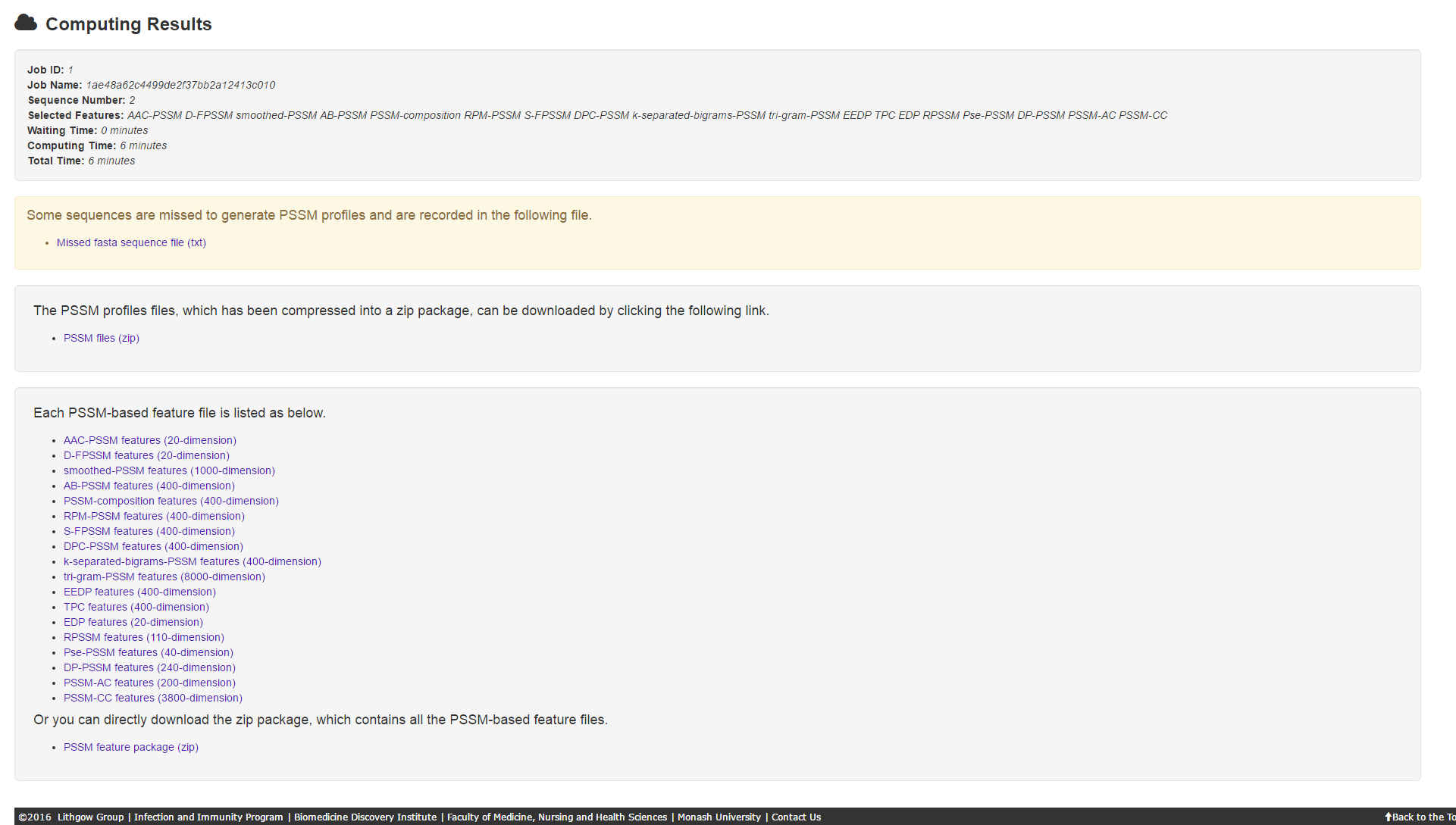
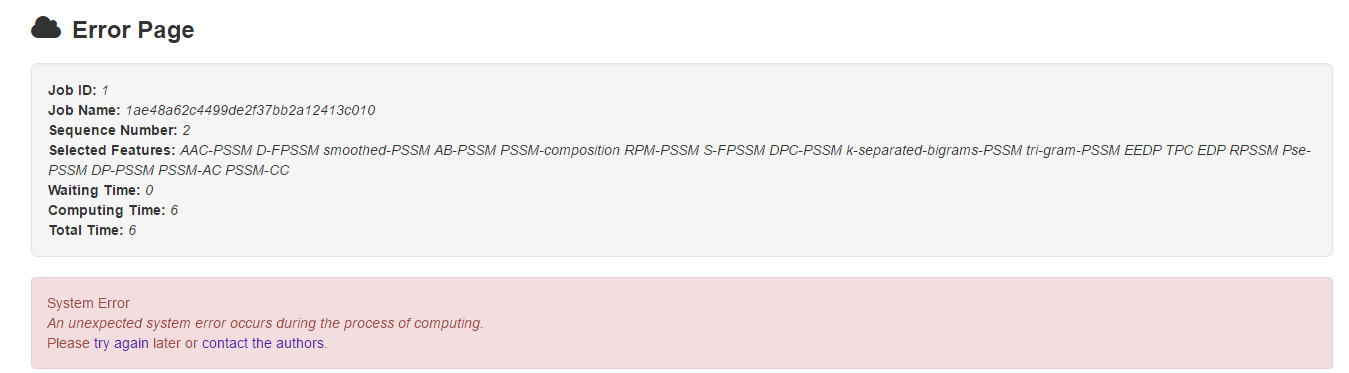
5.3 Error page
Should the computing process stop abnormally due to any unexpected system errors, users will get an error page which contains 2 parts: job information and error details.
STANDALONE SOFTWARE
1. Overview
The source codes of POSSUM can be downloaded at the DOWNLOAD page.
2. Using POSSUM
For users who prefer to apply their own parameter settings for specific research purposes and users who have the capacity to perform high throughput generation of PSSM files for a very large dataset using their local computers, an open source standalone software toolkit is also available. The standalone version of POSSUM was developed using Python and Perl, and can be executed on Unix/Linux, Windows and Mac OS. As an open source software, users can access, modify and redistribute the source codes, allowing users to tailor POSSUM according to their specific requirements.
2.1 System Requirements
-
Operating systems:
Windows,Unix/Linux,Mac OS - Dependencies:
2.2 File Description in the download directory
-
input: The input file folder (users can specify their own input file folder using `-i`).pssm_files: The PSSM file folder (users can specify their own PSSM file folder using `-p` ), which contains the example PSSM files.example_1.pssm, example_2.pssm: The example PSSM files.example.fasta: The example fasta file used to generate descriptors.
-
output: The folder used to store computational results of POSSUM (users can specify their own output folder using `-o`).*.csvfiles (such asexample_aac_pssm.csv,example_smoothed_pssm.csv,example_k_separated_bigrams_pssm.csv,example_pse_pssm.csv,example_dp_pssm.csv,example_pssm_ac.csv,example_pssm_cc.csv): The computational result files of the example fasta fileexample.fasta.
-
src: The source code folder.possum.py , possum_ft.py , featureGenerator.py , matrixTransformer.py: Python scripts used to generate raw descriptors.headerHandler.py: A Python script used to add headers for raw descriptors.
-
utils: The folder used to store a bunch of utility scripts that aiding users to formalize fasta sequences.removeIllegalSequences.pl: A Perl 5 script used to remove fasta sequences containing illegal characters, such as 'B', 'J', 'O', 'U', 'X' and 'Z'.
# usage examples: perl removeIllegalSequences.pl -i example.fasta -o example_corrected.fasta
removeShortSequences.pl: A Perl 5 script used to remove fasta sequences shorter than a given threshold value.
# usage examples: perl removeShortSequences.pl -i example.fasta -o example_corrected.fasta -n 50 perl removeShortSequences.pl -i example.fasta -o example_corrected.fasta -n 100
tmp: The folder used to cache temporary files in the process of program operation.
docs: The folder used to store help documents.
userguide.pdf: The detailed description file for POSSUM standalone toolkit.
possum_standalone.pl: A Perl 5 script facilating users to invoke and run POSSUM standalone toolkit.
2.3 Usage
Data preparation:
Two types of input files are needed for POSSUM:
-
fasta file: A fasta file should contain one/multiple protein sequences in fasta format. Users can specify a fasta file as input by using -i parameter. -
pssm_files: PSSM files for the fasta file (using BLAST against uniref 50/90/100 databases) should be provided in a certain folder, which will be specified by users using -p parameter.
Command line examples: For Unix/Linux/Mac OS X users:
perl possum_standalone.pl -i input/example.fasta -o output/example_aac_pssm.csv -t aac_pssm -p input/pssm_files -h T perl possum_standalone.pl -i input/example.fasta -o output/example_smoothed_pssm.csv -t smoothed_pssm -p input/pssm_files -h T -a 7 -b 50 perl possum_standalone.pl -i input/example.fasta -o output/example_k_separated_bigrams_pssm.csv -t k_separated_bigrams_pssm -p input/pssm_files -h T -a 1 perl possum_standalone.pl -i input/example.fasta -o output/example_pse_pssm.csv -t pse_pssm -p input/pssm_files -h T -a 1 perl possum_standalone.pl -i input/example.fasta -o output/example_dp_pssm.csv -t dp_pssm -p input/pssm_files -h T -a 5 perl possum_standalone.pl -i input/example.fasta -o output/example_pssm_ac.csv -t pssm_ac -p input/pssm_files -h T -a 10 perl possum_standalone.pl -i input/example.fasta -o output/example_pssm_cc.csv -t pssm_cc -p input/pssm_files -h T -a 10
For Windows users:
perl possum_standalone.pl -i input/example.fasta -o output/example_aac_pssm.csv -t aac_pssm -p input/pssm_files -h T perl possum_standalone.pl -i input/example.fasta -o output/example_smoothed_pssm.csv -t smoothed_pssm -p input/pssm_files -h T -a 7 -b 50 perl possum_standalone.pl -i input/example.fasta -o output/example_k_separated_bigrams_pssm.csv -t k_separated_bigrams_pssm -p input/pssm_files -h T -a 1 perl possum_standalone.pl -i input/example.fasta -o output/example_pse_pssm.csv -t pse_pssm -p input/pssm_files -h T -a 1 perl possum_standalone.pl -i input/example.fasta -o output/example_dp_pssm.csv -t dp_pssm -p input/pssm_files -h T -a 5 perl possum_standalone.pl -i input/example.fasta -o output/example_pssm_ac.csv -t pssm_ac -p input/pssm_files -h T -a 10 perl possum_standalone.pl -i input/example.fasta -o output/example_pssm_cc.csv -t pssm_cc -p input/pssm_files -h T -a 10Or
perl possum_standalone.pl -i input\example.fasta -o output\example_aac_pssm.csv -t aac_pssm -p input\pssm_files -h T perl possum_standalone.pl -i input\example.fasta -o output\example_smoothed_pssm.csv -t smoothed_pssm -p input\pssm_files -h T -a 7 -b 50 perl possum_standalone.pl -i input\example.fasta -o output\example_k_separated_bigrams_pssm.csv -t k_separated_bigrams_pssm -p input\pssm_files -h T -a 1 perl possum_standalone.pl -i input\example.fasta -o output\example_pse_pssm.csv -t pse_pssm -p input\pssm_files -h T -a 1 perl possum_standalone.pl -i input\example.fasta -o output\example_dp_pssm.csv -t dp_pssm -p input\pssm_files -h T -a 5 perl possum_standalone.pl -i input\example.fasta -o output\example_pssm_ac.csv -t pssm_ac -p input\pssm_files -h T -a 10 perl possum_standalone.pl -i input\example.fasta -o output\example_pssm_cc.csv -t pssm_cc -p input\pssm_files -h T -a 10Or
perl possum_standalone.pl -i input\\example.fasta -o output\\example_aac_pssm.csv -t aac_pssm -p input\\pssm_files -h T perl possum_standalone.pl -i input\\example.fasta -o output\\example_smoothed_pssm.csv -t smoothed_pssm -p input\\pssm_files -h T -a 7 -b 50 perl possum_standalone.pl -i input\\example.fasta -o output\\example_k_separated_bigrams_pssm.csv -t k_separated_bigrams_pssm -p input\\pssm_files -h T -a 1 perl possum_standalone.pl -i input\\example.fasta -o output\\example_pse_pssm.csv -t pse_pssm -p input\\pssm_files -h T -a 1 perl possum_standalone.pl -i input\\example.fasta -o output\\example_dp_pssm.csv -t dp_pssm -p input\\pssm_files -h T -a 5 perl possum_standalone.pl -i input\\example.fasta -o output\\example_pssm_ac.csv -t pssm_ac -p input\\pssm_files -h T -a 10 perl possum_standalone.pl -i input\\example.fasta -o output\\example_pssm_cc.csv -t pssm_cc -p input\\pssm_files -h T -a 10
NOTE: The main usage difference between Windows and other OS is the file path format. POSSUM allows /,\,\\ as path separators on windows in accordance with users’ habits.
Parameters:
-
-i: Specify the input file path of a file in fasta format. -
-o: Specify the output file path for the computational result. -
-t: Specify one of 21 algorithms to generate descriptors, including including aac_pssm, d_fpssm, smoothed_pssm, ab_pssm, pssm_composition, rpm_pssm, s_fpssm, dpc_pssm, k_separated_bigrams_pssm, tri_gram_pssm, eedp, tpc, edp, rpssm, pse_pssm, dp_pssm, pssm_ac, pssm_cc, aadp_pssm, aatp and medp. -
-p: Specify the PSSM file folder path. -
-h <T/F>: For adding header or not.Default = T.
For -i,-o and -p, absolute and relative paths are both allowed.
For smoothed-PSSM algorithm:
If you set -t as smoothed_pssm, the following parameter can be used to specify by our program:
-
-a: Specify the smoothing_window. The smoothing_window denotes the size of smoothing window and should be an odd number.Default = 7. -
-b: Specify the sliding_window. The sliding_window denotes the size of sliding window.Default = 50.
For k-separated-bigrams-PSSM algorithm:
If you set -t as k_separated_bigrams_pssm, the following parameter can be used to specify by our program:
-
-a: Specify the k. k denotes the distance between the amino acid positions.Default = 1.
For Pse-PSSM algorithm:
If you set -t as pse_pssm, the following parameter can be used to specify by our program:
-
-a: Specify the ξ. The ξ denotes the ξ most contiguous PSSM scores along the protein chain.Default = 1.
For DP-PSSM algorithm:
If you set -t as dp_pssm, the following parameter can be used to specify by our program:
-
-a: Specify the α. The α denotes α-th amino acid afterward.Default = 5.
For PSSM-AC algorithm:
If you set -t as pssm_ac, the following parameter can be used to specify by our program:
-
-a: Specify the LG. The LG denotes the maximum distance of two residues along the sequence.Default = 10.
For PSSM-CC algorithm:
If you set -t as pssm_cc, the following parameter can be used to specify by our program:
-
-a: Specify the LG. The LG denotes the maximum distance of two residues along the sequence.Default = 10.
For other algorithms:
If you set -t as aac_pssm, d_fpssm, ab_pssm, pssm_composition, rpm_pssm, s_fpssm, dpc_pssm, tri_gram_pssm, eedp, tpc, edp, rpssm, aadp_pssm, aatp or medp, you need not input other arguments.
2.4 Input file check
For the input file in fasta format, if there exsits sequence(s) shorter than 50 or containing illegal characters, such as B, J, O, U, X and Z, the program will exit and show corresponding tips.
Please refer to the output message, accordingly use the utility scripts in utils folder to dispose of the fasta sequences, and then try again.
2.5 Annotation of the computational results:
-
computational results are represented in
csvformat.